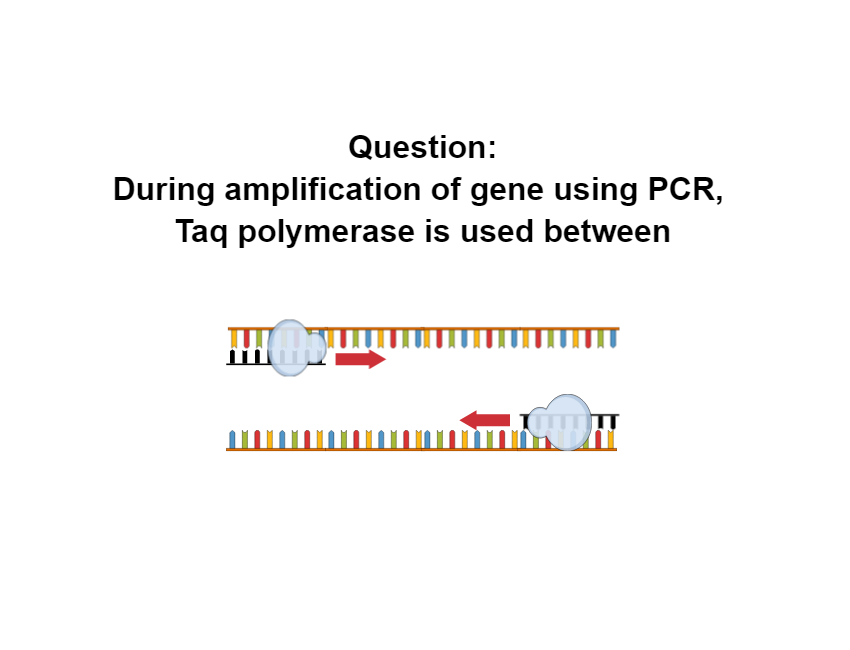
During amplification of gene using PCR, Taq polymerase is used between
[HDquiz quiz = “1967”] Polymerase chain reaction or PCR is a technique to amplify a specific segment of DNA using a pair of primers and […]

[HDquiz quiz = “1967”] Polymerase chain reaction or PCR is a technique to amplify a specific segment of DNA using a pair of primers and […]

[HDquiz quiz = “1954”] The genome of the first multicellular organism sequenced was Caenorhabditis elegans or C elegans. The genome of C elegans was sequenced […]

The Nirenberg and Matthaei experiment was performed by American biologist Marshall W. Nirenberg (1927-2010) and German biologist J. Heinrich Matthaei in 1961. Matthaeiat was a postdoc fellow in […]

The genome of E coli contains 4 million base pairs or 4×106 bp. The number of turns of DNA present is 400,000 turns or (4×105) […]
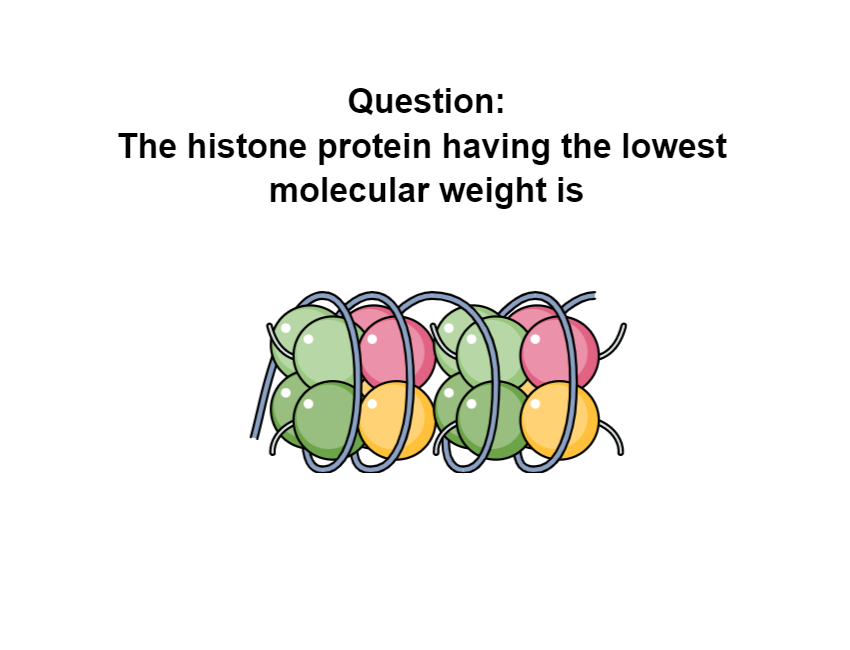
Histones are basic proteins that in association with DNA form a structure known as the nucleosome. Nucleosomes help in the condensation of DNA into chromatin […]

The bioinformatics tool that is not used to analyze the restriction enzymes sites of a DNA sequence is EuGene. NEBcutter, Webcutter, and WatCut are the […]

Erwin Chargaff proposed a rule called the Chargaff’s rules which states that in a double-stranded DNA segment, the ratio of purine bases (Adenine+Guanine) and the pyrimidine […]

DNA is tightly packed in the form of condensed chromatin structure to form chromosomes. The nucleosome is the unit of chromatin that form from eight […]

After transcription, a cap forms at the 5′ end of the mRNA in eukaryotes. The cap forms by the attachment of 7-methyl guanosine (m7G). 7-methyl […]

B-DNA is the right-handed DNA helix that is the original model of DNA proposed by JD Watson and F Crick. In the B-DNA model, two […]
Copyright © 2023 | WordPress Theme by MH Themes